Profiling 20k Immune Cells in Healthy PBMCs from a Single Encapsulation
- Rare cell types were identified in sample resulting from a single encapsulation, rather than from multiple merged datasets
- More than 20k cells were identified in T20 kit with > 50% capture
PBMCs obtained by Fluent BioSciences from AllCells (Lot 3082436) were used in a human cell experiment to demonstrate the robustness of PIPseq to identify heterogeneous immune cell types.
40K cells were input into a T20 PIPseq reaction, resulting in 23,779 captured cells. Libraries were generated using PIPseq v3.0 chemistry and sequenced on an Illumina NextSeq 2000 to a depth of 429,428,870 reads (~18K reads per cell). Paired-end dual indexing was performed with the following settings:
Read 1: 51 cycles, i5 index 8 cycles, i7 index 8 cycles, Read 2: 70 cycles
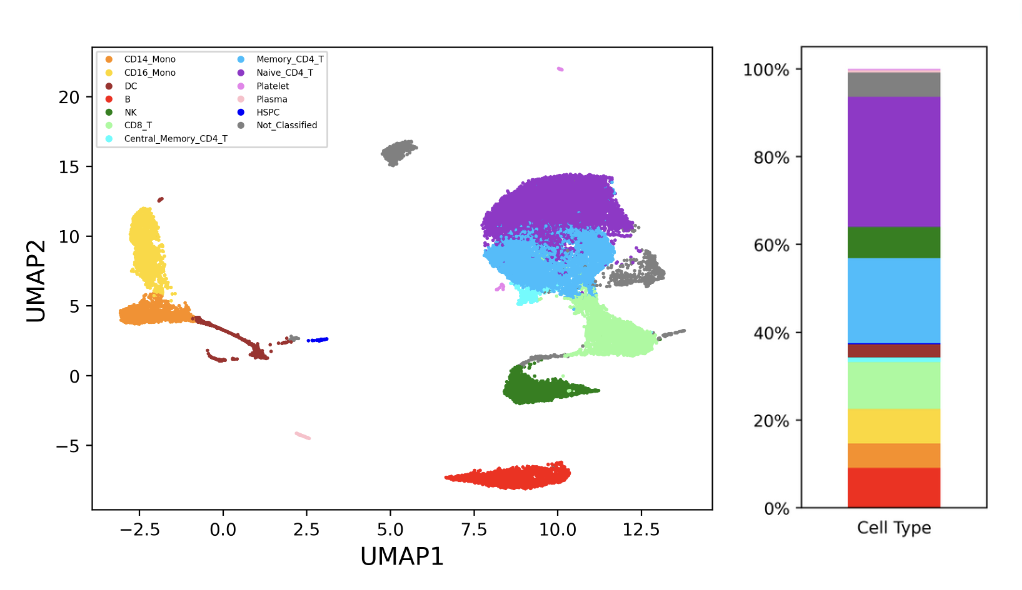
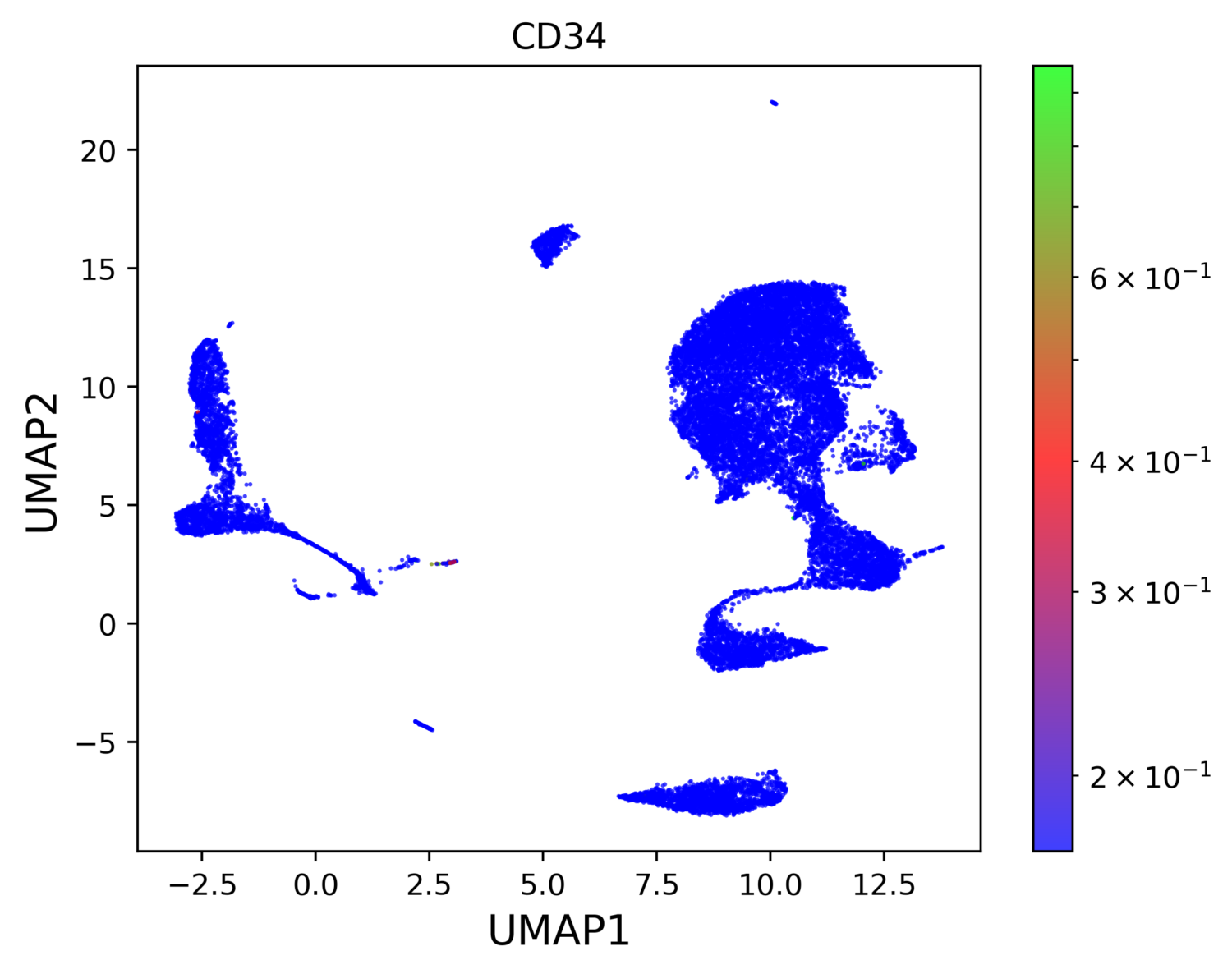
The sample was processed using PIPseeker v1.1.3 using default settings. Cell types were annotated by cluster (Figure 1). High resolution of the assay and large cell input allow for detection of many different cell populations. Common cell types (i.e., B, T, Monocytes), subtypes (e.g., Monocytes – CD14, CD16, DC), and rare cell populations (e.g., HSPCs) are all clearly resolved. Specifically, HSPCs are identified with only 56 cells (~0.23%), demonstrating the overall sensitivity of PIPseq. HSPCs show localized expression of canonical marker CD34 (Figure 2).
Resources
| File | Checksum |
|---|---|
| filtered_matrix.tar.gz | 29f452a6e1bo81d172c8ofc92d1aa047 |
| raw_matrix.tar.gz | 4b916fb39dc6fe32c24710b2a2g216a3 |
| starsolo_out.bam | a8a1d8bd99b00b35e1bd79accf0defa8 |
| fastqs.tar.gz | e297d6b57b469c677edadb2ce9534552 |
