SIngle Cell Analysis: Species Mixture Experiment with Large Cell Input
Clear Cluster Separation Demonstrates Low Doublet Rate
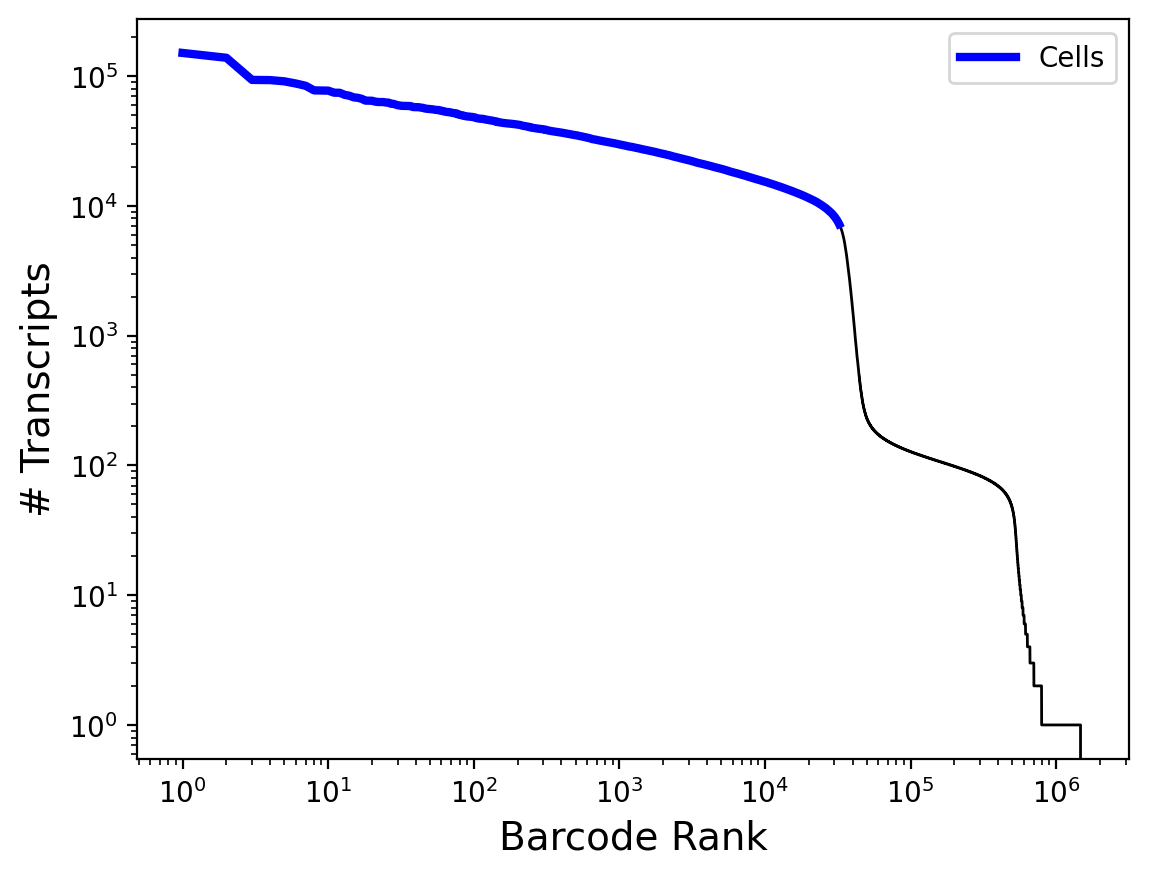
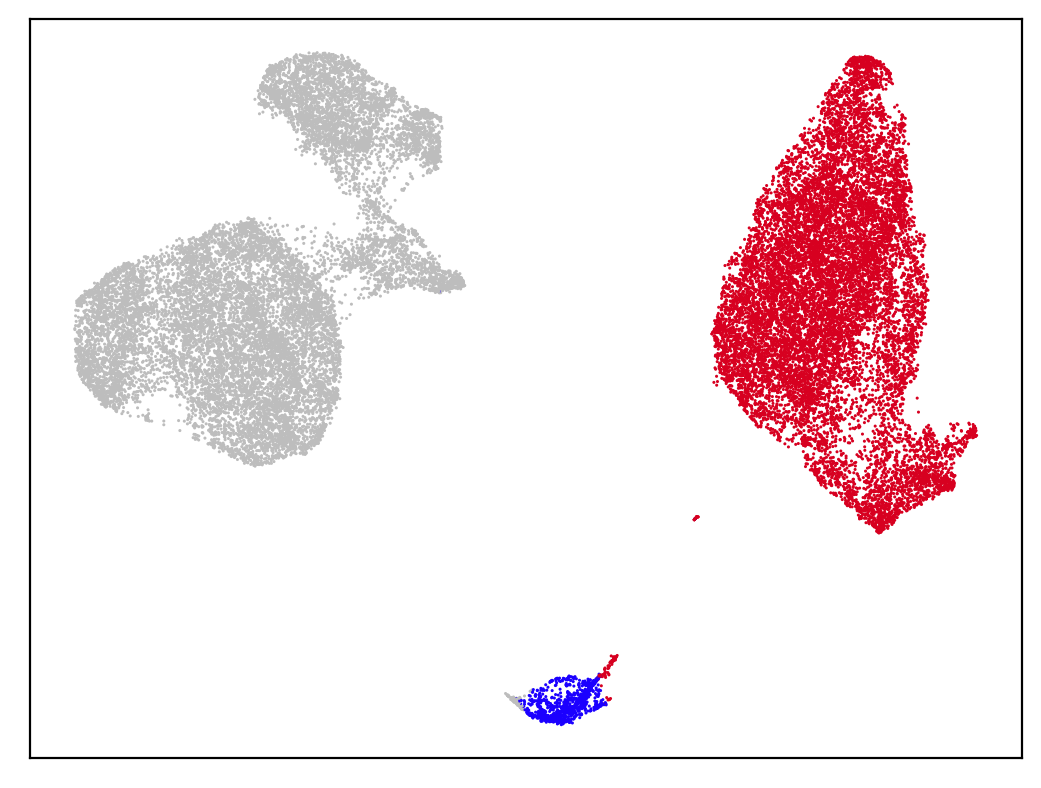
In this experiment, 39,440 cells from a 1:1 mixture of Human HEK 293T and Mouse NIH-3T3 were processed using the PIPseq v4 chemistry T20 3’ Single Cell RNA Kit. The goal of this experiment was to understand the maximum performance of the v4 chemistry and quantify standard “barnyard” metrics with regards to the chemistry and kit configuration. Samples were sequenced on an Illumina NextSeq 2000 sequencer to a depth of ~1.07 billion reads (~33k reads per cell). Paired-end dual indexing was used with the following settings:
- Read 1: 54 cycles
- Read 2: 67 cycles
- i5 index: 8 cycles
- i7 index: 8 cycles
Samples were processed using PIPseeker v3.0.0 using the flags –run-barnyard and –force-cells 32356. A UMAP dimensionality reduction projection shows clear separation between mouse and human cell populations, with a small cluster representing heterotypic multiplets (~3.3%).
