Mouse Brain Nuclei
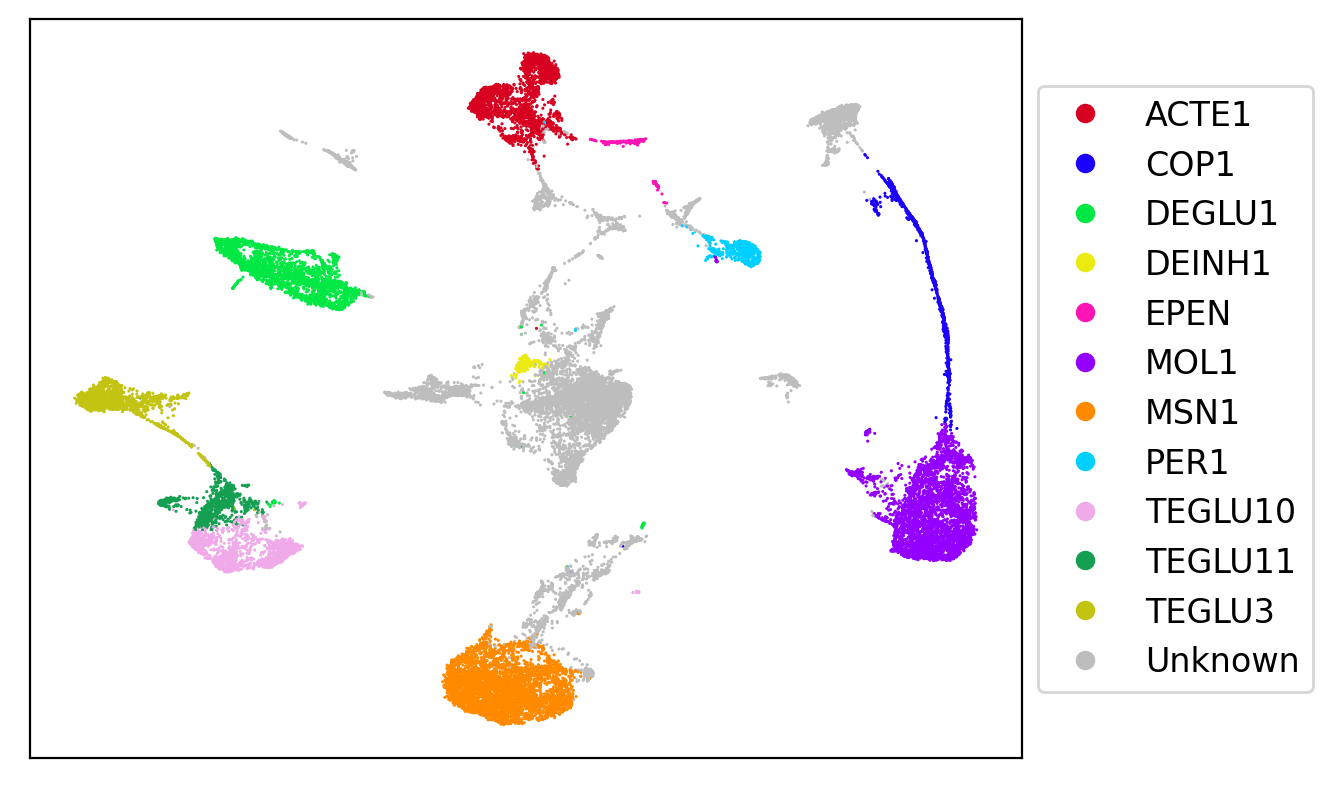
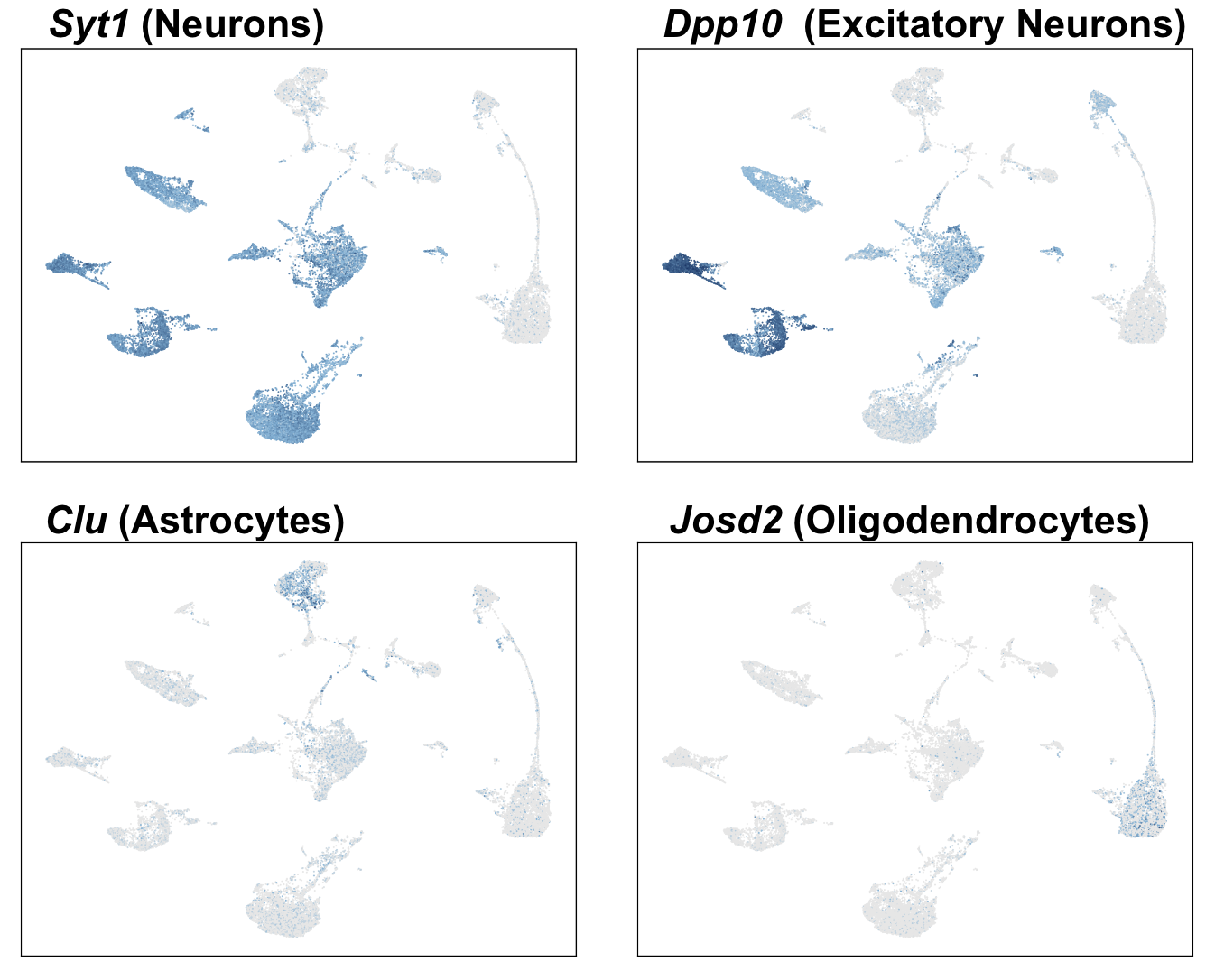
Cell types detected automatically with the whole brain annotation reference include: Astrocytes (ACTE1), Oligodendrocytes (COP1+MOL1), Excitatory Neurons (DEGLU1, TEGLU3,10,11), Inhibitory Neurons (DEINH1), Ependymal (EPEN), Medium Spiny Neurons (MSN1), and Pericytes (PER1). Gray-colored clusters remain unidentified; the annotation reference was derived from cells, which can have significantly different differential gene expression signatures from nuclei.
In this experiment, 40k nuclei were isolated using the PIPseq Nuclei Isolation Kit (FBS-SCR-NUC4) and input into a single tube of a PIPseq T100 V4 chemistry 3’ Single Cell RNA Kit. 30k nuclei were successfully captured. The goal of this experiment was to demonstrate the efficacy of the PIPseq nuclei isolation kit and compatibility of nuclei with PIPseq. The sample was sequenced on an Illumina NextSeq 2000 to a depth of ~387 million reads (~13K reads per cell). Paired-end dual indexing was used for both libraries with the following settings:
- Read 1: 54 cycles
- Read 2: 67 cycles
- i5 index: 8 cycles
- i7 index: 8 cycles
This sample was processed using PIPseeker v3.0.1. A diverse array of neurons, glia, vascular, and other neuronal cell types were identified.